qPCR Protocol for SNP Genotyping
实验概要
Platinum® qPCR SuperMix for SNP Genotyping is a ready-to-use reaction mix for the amplification and identification of single-nucleotide polymorphisms (SNPs) in genomic DNA using PCR-based SNP genotyping technologies such as fluorescent primers or probes. The SuperMix has been specifically formulated for discrimination of alleles by real-time qPCR or end-point PCR followed by allelic-discrimination analysis on a real-time instrument or fluorescent microplate reader. It provides enhanced fluorescent signals for better discrimination of alleles and good separation with minimal scattering between replicate samples. The SuperMix format and integrated UDG carryover prevention make this reagent well suited for high-throughput applications.
This SuperMix has been developed using Applied Biosystems’ TaqMan®-based SNP genotyping products.
主要试剂
Platinum® qPCR SuperMix for SNP Genotyping [Details:contains Platinum® Taq DNA polymerase, Tris-HCl (pH 8.4), 6 mM MgCl2, 400 µM dGTP, 400 µM dATP, 400 µM dCTP, 800 µM dUTP, uracil DNA glycosylase (UDG), and PCR enhancer and proprietary stabilizers]
实验步骤
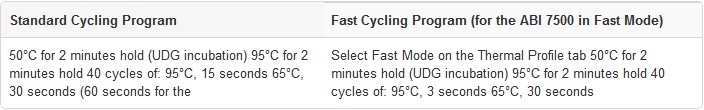
1. Program the real-time instrument or standard thermal cycler to perform a brief UDG incubation immediately followed by PCR amplification, as shown below. Optimal cycling temperatures and times may vary for different target sequences and primer/probe sets.
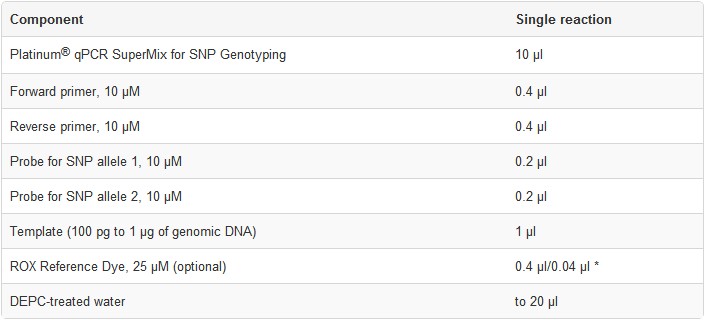
2. Prepare each reaction in a microcentrifuge tube or PCR plate wellas specified below.For multiple reactions, prepare a master mix of common components, add the appropriate volume to each tube or plate well, and then add the unique reaction components (e.g., template).
Note: Preparation of a master mix is essential in quantitative applications to reduce pipetting errors.
3. Cap or seal the reaction tube/PCR plate, and gently mix. Make sure that all components are at the bottom of the tube/plate; centrifuge briefly if needed.
4. Place reactions in the thermal cycler programmed as described above and run the program.
5. For real-time instruments, perform real-time analysis and/or an allelic-discrimination end-point reading at the end of the run. For standard thermal cyclers, transfer the PCR product to a fluorescent microplate reader for SNP genotyping analysis.
注意事项
1. The protocol uses TaqMan® probes in a SNP genotyping assay on ABI real-time instruments or a standard thermal cycler. Note the separate cycling conditions for the ABI 7500 in Fast Mode, and the lower amount of ROX Reference Dye required for the ABI 7500 and 7500 Fast systems. This generic protocol may also be suitable with some modifications for other real-time instruments.
2. For additional information about specific instruments click here . Since PCR is a powerful technique capable of amplifying trace amounts of DNA, all appropriate precautions should be taken to avoid cross-contamination.
3. The target template for SNP genotyping is purified genomic DNA. For a 20-µl reaction, use 10 ng to 1 µg of purified genomic DNA in a 1µl volume. To purify genomic DNA, we recommend the PureLink™ or ChargeSwitch® genomic DNA purification kits from Invitrogen.
4. Platinum®qPCR SuperMix for SNP Genotyping includes magnesium chloride at a final concentration of 3 mM, which is optimal for SNP genotyping experiments.
5. ROX Reference Dye is included in each kit to normalize the fluorescent reporter signal for instruments that are compatible with this option. ROX is supplied at a 25 µM concentration, and is composed of a glycine conjugate of 5-carboxy-X-rhodamine, succinimidyl ester in 20 mM Tris-HCl (pH 8.4), 0.1 mM EDTA, and 0.01% Tween® 20. Use the following table to determine the amount of ROX to use with a particular real-time instrument per 20-µl reaction:

6. Platinum® qPCR SuperMix for SNP Genotyping can be used with real-time qPCR instruments that can detect three colors (one for each SNP, plus one for ROX Reference Dye). Supported real-time instruments include the ABI PRISM® 7000, 7700, and 7900HT; the ABI 7300 and 7500 Real-Time PCR Systems; the Stratagene Mx3000P®, Mx3005P™, and Mx4000® the Corbett Research Rotor-Gene™; and the MJ Research DNA Engine Opticon® and Opticon® 2.
You can also perform end-point PCR on a standard thermal cycler and analyze the results using a fluorescent microplate reader capable of detecting in three channels.
7. SNP genotyping using fluorescent dual-labeled probe technology such as TaqMan® probes requires two PCR primers as well as two allele-specific probes that hybridize to the SNP portion of the amplicon. Predeveloped TaqMan® SNP genotyping assays are available from Applied Biosystems Assays-on-Demand, and include a mix of PCR primers and two allele-specific TaqMan®MGB Probes, labeled with FAM and VIC. Custom TaqMan® SNP genotyping probes labeled with FAM and VIC may be designed using Applied Biosystems’ Primer Express 2.0 software.
When designing custom probes, note that the probe sequences should be free of secondary structure and should not hybridize to each other or to primer 3´-ends. The optimal concentration of the probes may vary between 50 and 800 nM, with a recommended starting concentration of 100 nM each.
PCR primers used with custom probes should be designed according to standard PCR guidelines. They should be specific for the target sequence and free of internal secondary structure, and should avoid complementation at 3´-ends within each primer, with each other, or with the dual-labeled probes. Optimal results may require a titration of primer concentrations between 100 and 500 nM. A final concentration of 200 nM per primer is effective for most reactions.
附 件 (共3个附件,占110KB)
![]()
图三.jpg
37KB 查看![]()
图二.jpg
41KB 查看![]()
图一.jpg
32KB 查看











