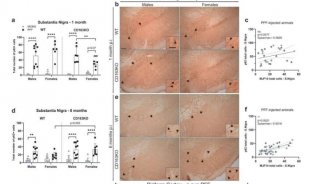
Inhibition of Huntington's disease neurodegeneration

Huntington's disease is a neurodegenerative condition caused by a dominant mutation in a gene encoding a protein now called huntingtin. Large polyglutamine repeats in the huntingtin protein are the genetic defect responsible for this condition, caused by expansion of a three-base pair repeat in the gene. Similar expansions of polyglutamine repeats are also found in other genes involved in neurodegenerative conditions. The polyglutamine repeat makes huntingtin protein insoluble and triggers the formation of protein aggregates. There are a few hypotheses for the connection between these aggregates and the neurodegeneration observed in Huntington's disease. One hypothesis is that mutant huntingtin moves into the nucleus and interferes with transcriptional activation involving the transcriptional coactivator CBP (CREB binding protein) and other coactivators. CBP interacts with transcriptional regulatory factors and with the basal transcriptional machinery to help regulate transcription. CBP is a large protein with multiple domains and in addition to interacting with several different transcription factors also has a histone acetylase enzyme activity. Mutant huntingtin binds to CBP, drawing it into insoluble protein aggregates. Mutant huntingtin also interacts directly with the acetyltransferase domain of CBP, blocking this activity. Activation of CBP acetyltransferase activity by transcriptional regulators results in the acetylation of histones in the promoter and enhancer regions of active genes, contributing to transcriptional activation by making these genes more accessible in chromatin. The lack of CBP in Huntington's affected neurons may lead to transcriptional repression of key genes that in turn leads to neurodegeneration. One potential treatment being tested is the treatment of Huntington's affected individuals with histone deacetylase inhibitors to restore normal acetylation levels and transcription. The repression of transcription may also be involved in other neurogenerative conditions caused by proteins with expansions of polyglutamine repeats.
Contributor:
REFERENCES: Karpuj MV, et al. Prolonged survival and decreased abnormal movements in transgenic model of Huntington disease, with administration of the transglutaminase inhibitor cystamine. Nat Med, vol 8(2), February 2002, 143-49. Kegel, K.B., et al. Huntingtin is present in the nucleus, interacts with the transcriptional corepressor C-terminal binding protein, and represses transcription. Biochem. J., vol 277(9), March 2002, 7466-76. Kim, Yun, et al. Caspase 3-cleaved N-terminal fragments of wild-type and mutant huntingtin are present in normal and Huntingtons disease brains, associate with membranes, and undergo calpain-dependent proteolysis. PNAS, vol 98(22), October 2001, 12784-89. McCampbell, A., et al. Histone deacetylase inhibitors reduce polyglutamine toxicity. PNAS, vol 98(26), 2001, 15179-84. Neri Christina. New light on polyglutamine neurodegenerative disorders: interference with transcription. TRENDS Mol. Med, vol 7(7), July 2001, 283-84. Nucifora, F.C.Jr., et al. Interference by huntingtin and atrophin-1 with cbp-mediated transcription leading to cellular toxicity. Science, vol 291(5512), March 2001, 2423-48. Steffan, J.S. The Huntingtons disease protein interacts with p53 and CREB-binding protein and represses transcription. PNAS, vol 97(12), 2000, 6763-68. Steffan, J.S., et al. Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila. Nature, vol 413(6857), October 2001, 739-43. Wyttenbach, Andreas, et al. Polyglutamine expansions cause decreased CRE-mediated transcription and early gene expression changes prior to cell death in an inducible cell model of Huntingtons disease. Hum. Mol. Genet., vol 10(17), 2001, 1829-45.