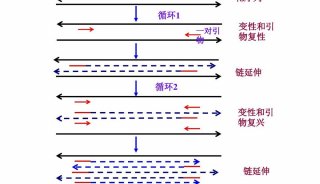
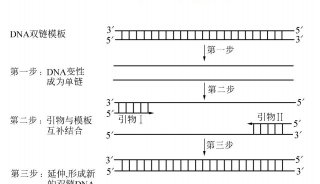
PCR Primer Design(三)
References
Albert, J., and Fenyo, E.M. 1990.
Simple, sensitive and specific detection of human immunodeficiency virus
type 1 in clinical specimens by polymerase chain reactions with nested
primers. J. Clin. Microbiol. 28: 1560-1564.
Berg, E.S. and
Olaisen, B. 1994. Hybrid PCR sequencing: sequencing of PCR products
using a universal primer. Biotechniques 17: 896-901.
Bindon,
C., Martindale, J., and Mitchell, C. 1998. Biologically generated primer
for PCR: PCR primer for unknown sequence. Nucleic Acid Res. 26(13):
3305-3358.
Breslauer, K.J., Ronald, F., Blocker, H., and
Marky, L.A.1986. Predicting DNA duplex stability from the base sequence.
Proc. Natl. Acad. Sci. 83: 3746-3750.
Bulat, S.A., Kobaev,
O.K., Mironenko, N.V., Ibatullin, F.M., Luchkina, L.A., and Suslov, A.V.
1992. Polymerase chain reaction with universal primers for studying
genomes. Genetika 28: 19-28.
Chamberlain, J.S., Gibbs, R.A.,
Ranier, J.E., and Caskey, C.T. 1991. Detection of gene deletions using
multiplex polymerase chain reactions, Meth. Molec. Biol. 9: 299-312.
Chang,
J.G., Lu, J.M., Huang, J., Chen, J.T., Liu, H.J., and Chang, C.P. 1995.
Rapid diagnosis of b-thalassaemia by mutagenically separated polymerase
chain reaction (MS-PCR) and its application to prenatal diagnosis, Br.
J. Haemat. 91: 602-605.
Chen, H., and Zhu,G.1997. Computer
program for calculating the melting temperature of degenerate
oligonucleotides used in PCR or hybridization. Biotechniques 21: 134-140
Chou, Q., Russell, M., Birch, D.E., Raymond, J., and Bloch,
W. 1992. Prevention of pre-PCR mis-priming and primer dimerization
improves low-copy number amplifications. Nucleic Acids Res. 20:
1717-1723.
Dieffenbach, C.W., Lowe, T.M.J., and Dveksler, G.S.
1993. General concepts for PCR primer design. In: PCR Methods and
Applications, Cold Spring Harbor Laboratory 3: S30-S37.
Drummond,
R., and Gelfand, D.H. 1989. Isolation, characterization and expression
in Escherichia coli of the DNA polymerase gene from Thermus aquaticus.
J. Biol. Chem. 264: 6427-6436.
Edwards, M.C., and Gibbs, R.A. 1994. Multiplex PCR: Advantages, development and applications. PCR Methods Applic. 3: S65-S75.
Engelke,
D.R., Hoener, P.A., and Collins, F.S. 1988. Direct sequencing of
enzymatically amplified human genomic DNA. Proc. Natl. Acad. Sci. 85:
544-550.
Erlich, H.A., Gefland, D., and Sninsky, J.J. 1991. Recent advances in the polymerase chain reaction. Science 252: 1643-1651.
Freier,
S.M., Kierzek, R., Jaeger, J.A., Sugimoto, N., Caruthers, M.H.,
Neilson, T., and Turner, D.H. 1986. Improved free-energy parameters for
predictions of RNA duplex stability. Proc. Natl. Acad. Sci, USA. 83:
9373-9377.
Green, C.E., Lund, J.K., and Manos, M.M. 1991. PCR
amplification from parafin-embedded tissues: Recommendations on
fixatives for long term storage and prospective studies. PCR Methods
Appl. 1: 46-50.
Haff La. 1994. Improved quantitative PCR using nested primers. PCR Methods Appl. 3: 322-327.
He,
Q., Marjamaki, M., Soini, H., Mertsola, J., and Viljanen, M.K. 1994.
Primers are decisive for sensitivity of PCR. Biotechniques. 17: 82-87.
Huang,
M., Arnheim, N., and Goodman, M.F. 1992. Extension of base mispairs by
Taq DNA polymerase: Implications for single nucleotide discrimination in
PCR. Nucleic Acids Res. 20: 4567-4573.
Hung, T., Mak, K. and Fong, K. 1990. A specificity enhancer for PCR, Nucleic Acids Res. 18: 4953-4959.
Jung,
V., Pestka, S.B., and Pestka, S. 1990. Efficient cloning of PCR
generated DNA containing terminal restriction endonuclease sites.
Nucleic Acid Res. 18: 6156-6160.
Kain, K.C., Orlandi, P.A.,
and Lanar, D.E. 1991. Universal promoter for gene expression without
cloning Expression-PCR. BioTechniques 10: 366-374.
Kaneko, S.,
Feinstone, S.M., and Miller, R.H. 1989. Rapid and sensitive method for
the detection of serum hepatitis B virus DNA using the polymerase chain
reaction technique. J. Clin. Microbiol. 27: 1930-1933.
Kawasaki,
E.S. 1990a. Sample preparation from blood, cells and other fluids, In:
PCR Protocols, a guide to methods and applications, Innis, M.A.,
Gefland, D.H., Sninsky, J.J. and White, T.J. (eds), Academic Press Inc.,
Berkeley CA, pp. 146-152.
Kawasaki, E.S. 1990b. Amplification
of RNA, In: PCR Protocols, a guide to methods and applications, Innis,
M.A., Gefland, D.H., Sninsky, J.J. and White, T.J. (eds), Academic Press
Inc., Berkeley CA, pp. 21-27.
Keller, G.H., Cumming, C.U.,
Huang, D.P., Manak, M.M., Ting, R. 1988. A chemical method for
introducing Haptens onto DNA probes. Anal. Biochem. 170: 441-450.
Klebe,
R.J., Grant G.M., Grant, A.M., Garcia, M.A., Giambernardi, T.A., and
Taylor, G.P. 1996. RT-PCR without RNA isolation. Biotechniques 21:
1094-1100.
Kumar A., and Mishra B.N. 1997. Primer Premier 4. Biotech Software and Internet Journal 3: 31-38.
Kwok
S., Kellogg D.E., McKinney, E., Spasic, D., Levenson, C., Sninsky, J.J.
1990. Effects of Primer-template mismatches on the polymerase chain
reaction: Human Immunodeficiency virus type I model studies, Nucleic
Acids Res. 18: 999-1005.
Kwok, S., Chang, S.Y., Sninsky, J.J.,
and Wong, A. 1994. A guide to the design and use of Mismatched and
Degenerate primers. PCR Methods and Appl. S539-S547.
Lawyer,
F.C., Stoffel, S., Saiki, R.K., Myambo, K., Drummond, R., and Gelfand,
D.H. 1989. Isolation, characterization, and expression in Escherichia
coli of the DNA polymerase gene from Thermus aquaticus. J. Biol. Chem.
15: 6427-6437.
Le Guyader, F., Estes, M.K., Hardy, E., Neill,
F.H., Green, J., Brown, D.W., and Atmar, R.L. 1996. Evaluation of a
degenerate primer for the PCR detection of human caliciviruses. Arch.
Virol. 141: 2225-2235.
Liang, P., and Pardee, A. 1992. Differential display of eukaryotic mRNAs by PCR. Science 257: 967-971.
Linz,
U., Delling, U., and Rubsamem-Waigmann 1990. Systematic studies on
parameters influencing the performance of the polymerase chain reaction.
J. Clin. Chem. Clin. Biochem. 28: 5-13.
Liu, Q., Thorland
E.C., and Sommer, S.S. 1997. Inhibition of PCR amplification by a point
mutation down stream of a primer. Biotechniques 22: 292-298.
Lowe,
T., Shareifkin, J., Yang, S.Q., and Dieffenbach, C.W. 1990. A computer
program for selection of oligonucleotide primers for polymerase chain
reaction. Nucleic Acids Res. 18: 1757-1761.
Lucas, K.M.,
Busch, S.M., and Thompson, J.A. 1991. An improved microcomputer program
for finding gene or gene family-specific oligonucleotides suitable as
primers for polymerase chain reactions or as probes. CABIOS 7: 525-529.
Mack,
D.H., and Sninsky, J.J. 1988. A sensitive method for the identification
of uncharacterized viruses related to known virus groups: Hepadnavirus
model system. Proc. Natl. Acad. Sci. 85: 6977-6981.
Makarova,
K.S., Mazin, A.V., Wolf, Y.I., and Soloviev, V.V. 1992. DIROM: An
experimental design interactive system for directed mutagenesis and
nucleic acid engineering. CABIOS 8: 425-431.
Manos, M.M.,
Ting, Y., Wright, D.K., Lewis, A.J., Broker, T.R., and Wolinsky, S.M.
1989. Use of polymerase chain reaction amplification for the detection
of genital human papillomaviruses. Cancer Cells (Molecular diagnostics
of Human Cancer) 7: 209-214.
Marky, L.A., Blumenfeld, K.S.,
Kozlowski, S., and Breslauer, K.J. 1983. Salt-dependent conformational
transitions in the self-complementary deoxydodecanucleotide
(CGCAATTCGCG): evidence for hairpin formation. Biopolymers 22: 1247-57.
Marky,
L.A., Kallenbach, N.R., Seeman, N.C., Haasnoot, C.A.G., and Breslauer,
K.J., 1985. Fourth Conversation in Biomolecular stereodynamics, Ed.
Sharma R.H. Adenine Guilderland, NY.
Mercier, B., Gaucher, C.,
Feugeeas, O., Mazurier, C. 1990. Direct PCR from whole blood without
DNA extraction. Nucleic Acid Res. 18(19): 5908-5912.
Moore,
R.E., Shepherd, J.W., and Hoskins, J. 1990. Design of PCR primers that
detect only mRNA in the presence of DNA, Nucleic Acids Res. 18:
1921-1926.
Mullis, K.B., and Faloona, F. 1987. Specific
synthesis of DNA in vitro via a polymerase-catalyzed chain reaction.
Methods Enzymol. 155: 335-351.
Murray, M., and Thompson, W. 1980. Rapid isolation of high molecular weight plant DNA, Nucleic Acids Res. 8: 4321-4325.
Nagano,
I., Zhiling, W., Nakayama, M., Takahashi, Y. 1996. A simple method to
design PCR primer to detect genomic DNA of parasites and its application
to Diorofilaria immitis. Mol. Cell. Probes. 10: 423-425.
Nelson,
J.W., Martin, F.H., and Tinoco, I., Jr. 1981. DNA and RNA oligomer
thermodynamics: the effect of mismatched bases on double-helix
stability. Biopolymers. 20: 2509-2531.
Newton, C.R., Graham,
A., Heptinstall, L.E., Powell, S.J., Summers, C., Kalsheker, N., Smith,
J.C., and Markham, A.F. 1989. Analysis of any point mutation in DNA: The
amplification refractory mutation system (ARMS). Nucleic Acid Res. 17:
2503-2516.
Nicodeme, P. and Steyaert J.M. 1997. Selecting optimal oligonucleotide primers for multiplex PCR ISMB 5:210-213.
Nomenclature
Committee of the International Union of Biochemistry 1985. Nomenclature
for incompletely specified bases in nucleic acid sequences. Eur. J.
Biochem. 150: 1-5.
O'Hara, P.J., and Venezia, D. 1991. PRIMGEN: a tool for designing primers from multiple alignments. CABIOS 7: 533-534.
Old, J.M., 1991. Detection of Mutations by the Amplification Refractory Mutation System (ARMS), Meth. Mol. Biol. 9: 77-84.
Osborne,
B.I. 1992. Hyper PCR: A Macintosh Hypercard program for determination
of optimal PCR annealing temperature. CABIOS 8: 33-34.
Patel,
D.J., Kozlowski, S.A., Marky, L.A., Rice, J.A., Broka, C., Itakura, K.,
and Breslauer, K.J. 1982. Structure and energetics of a hexanucleotide
duplex with stacked trinucleotide ends formed by the sequenced
(GAATTCGCG). Biochemistry. 21: 451-455.
Patel, D.J.,
Kozlowski, S.A., Marky, L.A., Rice, J.A., Broka, C., Itakura, K., and
Breslauer, K.J. 1983. Topics in Nucleic acid structure, Ed. Neidle, S.
Wiley, NY. pp.81-136.
Petruska, J., Goodman, M.F., Boosalis,
M.S., Sowers, L.C., Cheong, C., Tinoco, I.Jr. 1988. Comparison between
DNA melting thermodynamics and DNA polymerase fidelity. Proc. Natl.
Acad. Sci. 85: 6252-6256.
Plasterer, T.N. 1997. Primerselect: Primer and probe design. Methods Mol. Biol. 70: 520-527.
Ponce, M.R., and Micol, J.L. 1992. PCR amplification of long DNA fragments, Nucleic Acids Res. 20: 623-627.
Rose,
T.M., Schultz, E.R., Henikoff, J.G., Peitrokovski, S., McCallum, C.M.,
and Henikoff, S. 1998. Consensus degenerate hybrid oligonucleotide
primers for amplification of distantly related sequences. Nucleic Acid
Res. 26: 1628-1635.
Roux, K.H. 1995. Optimization and
troubleshooting in PCR in: PCR methods and applications. Cold spring
Harbor laboratory. 5185-5194.
Rychlik, W., and Rhoads, R.E.
1989. A computer program for choosing optimal oligonucleotides for
filter hybridization, sequencing and in vitro amplification of DNA.
Nucleic Acids Res. 17: 8543-8551.
Rychlik, W., Spencer. W.J.,
and Rhoads, R.E. 1990. Optimisation of annealing temperature for DNA
amplification in vitro. Nucleic Acids Res. 18: 6409-6412.
Saiki,
R.K., Scharf, S., Faloona, F., Mullis, K.B., Horn, G.T., Erlich, H.A.,
and Arnheim, N. 1985. Enzymatic amplification of beta-globin genomic
sequences and restriction site analysis for diagnosis of sickle cell
anemia. Science 230: 1350-1354.
Sambrook, J., Fritsch, E.F.,
and Maniatis, T. 1989. Molecular Cloning, A laboratory manual, 2nd Ed.,
Cold Spring Harbor Laboratory Press.
SantaLucia, J.Jr.,
Allawi, H.T. and Seneviratne, P.A. 1996. Improved Nearest-Neighbor
parameters for predicting DNA duplex stability. Biochemistry 35:
3555-3562.
Sarkar, G., Kapelner, S., and Sommer, S.S. 1990.
Formamide can dramatically improve the specificity of PCR. Nucleic Acids
Res. 18: 7465-7468.
Schowalter, D.B., and Sommer, S.S. 1989.
The generation of radiolabelled DNA and RNA probes with polymerase chain
reaction, Anal. Biochem. 177: 90-94.
Sheffield, V.C., Cox,
D.R., Lerman, L.S., and Myers, R.M. 1989. Attachment of a 40 base pair
G+C rich sequence (GC-clamp) to genomic DNA fragments by polymerase
chain reaction results in improved detection of single base changes.
Proc. Natl. Acad. Sci. 86: 232-236.
Shuber, A., Skoletsky, J.,
Handelin, B. 1991. A rapid method for simultaneous analysis of multiple
point mutations within the Cystic Fibrosis gene. Am. J. Hum. Genet. 49:
189-194.
Singh, V.K., Mangalam, A.K., Dwivedi, S., and Naik, S.
1998. Primer Premier: Program for design of degenerate primers from a
protein sequence, Biotechniques 24: 318-319.
Singh V.K., and
Naik, S. 2000. Room temperature stable polymerase chain reaction kit for
detection of Hepatitis B Virus. Clin. Chim. Acta. 301: 225-227.
Singh,
V.K., Govindarajan, R., Naik, S., and Kumar, A. 2000. The effect of
hairpin structure on PCR Amplificaiton efficiency. Mol. Biol. Today. 1:
657-659.
Singh, V.K., Manglam, A.K., Kumar, A., and Naik, S.
2001. Universal primers can amplify tumour necrosis factor gene across
species. Mol. Biol. Today 2: 11-12.
Singh V.K., and Naik, S.
2001. Simultaneous amplification of DNA and RNA virus using Multiplex
PCR system. Clin. Chim. Acta. (In Press).
Sze, S.H., Raytberg,
M.A., Gelfand, M.S., Mironov, A.A., Astakhova, T.V., Pevzner, P.A.
1998. Algorithms and software for support of gene identification
experiments. Bioinformatics 14: 14-19.
Tamura, T., Holbrook,
S.R., and Kim, S.H. 1991. A Macintosh computer program for designing DNA
sequences that for specific peptides and proteins. BioTechniques. 10:
782-784.
Tan, J.A.M.A., Tay, J.S.H., Lin, L.I., Kham, S.K.Y.,
Chia, J.N., Chin, T.M., Norkamov, B.T., Aziz, A.O.B., and Wong, H.B.
1994. The amplification refractory mutation system (ARMS): a rapid and
direct prenatal diagnostic technique for b-thalassaemia in Singapore.
Prenat. Diagn. 14: 1077.
Tinoco, I.Jr., Borer, P.N., Dengler,
B., Levin, M.D., Uhlenbeck, O.C., Crothers, D.M., and Bralla, J. 1973.
Improved estimation of secondary structure in ribonucleic acids. Nat.
New Biol. 14: 40-41.
Tinoco, I.Jr., Uhlenbeck, O.C., and Levine,
M.D. 1971. Estimation of secondary structure in ribonucleic acids.
Nature 9: 362-367.
Uhlenbeck, O.C., Borer, P.N., Dengler, B.,
and Tinoco, I.Jr. 1973. Stability of RNA hairpin loops: A 6 -C m -U 6.
J. Mol. Biol. 5: 483-496.
Varawall, N.Y., Old, J.M., and
Weatherall, D.J. 1991b. Rare b-thalassaemia mutations in Asian Indians,
Br. J. Haemat. 79: 640-644.
Varawalla, N.Y., Old, J.M.,
Sarkar, R., Venkatesan, R., and Weatherall, D.J. 1991a. The spectrum of
b-thalassaemia mutations on the Indian subcontinent: the basis for
prenatal diagnosis, Br. J. Haemat. 78: 242-247.
Venta, P.I.,
Brouillette, J.A., Yusbasiyan-Gurkan, V., and Brewer, G.J. 1996. Gene
specific universal mammalian sequence ? tagged sites: application to the
canine genome. Biochem. Genet. 34: 321-341.
Wilks, F.F.,
Kurban, R.R., Hovens, C.M., and Ralph, S.J. 1989. The application of the
polymerase chain reaction to cloning members of the protein tyrosine
kinase family. Gene. 85: 67-74.
Wilson, J.T., Wilson, L.B.,
deRiel, J.K., Villa-Komaroff, L., Efstratiadis, A., Forget, B.G., and
Weissman, S.M. 1978. Insertion of synthetic copies of human globin genes
into bacterial plasmids. Nucleic Acids Res. 5: 563-581.
Wrischnick,
L.A., Higuchi, R.G., Stoneking, M., Erlich, H.A., Arnheim, N., and
Wilson, A.C. 1987. Length mutations in human mitochondrial DNA: Direct
sequencing of enzymatically amplified DNA. Nucleic Acids Res. 15:
529-542.
Wu, D.Y.W., Ugozzoli, B.K., Pal, J., Qian, and
Wallace, R.B. 1991. The effect of temperature and oligonucleotide primer
length on specificity and efficiency of amplification by the polymerase
chain reaction. DNA Cell Biol. 10: 233-238.
Zhu, K.Y., and
Clark, J.M. 1996. Addition of competitive primer can dramatically
improve the specificity of PCR amplification of specific alleles.
Biotechniques. 21: 586-590.