E.Z.N.A.® Total RNA Maxi Kit Protocol for Animal Tissues
实验概要
The E.Z.N.A.® Total RNA Maxi Kit uses HiBind® matrix spin-column technology to isolate up to 5 mg total cellular RNA from a variety of sources including 1010 bacterial cells, 1 g tissue, and up to 5 x 108 cultured cells. Samples are lysed and homogenized in denaturing conditions to ensure degradation of proteins and inactivation of endogenous RNases. Binding conditions are then optimized to favor RNA isolation before the resulting cell lysate are passed through HiBind® RNA Maxi spin-columns. Subsequent wash steps remove cellular debris and trace salt contaminants to allow elution of pure RNA in DEPC-treated dH2O. No organic solvents are used and only short centrifugation steps using a standard centrifuge equipped with a swinging-bucket rotor at room temperature are needed. The kit can also be used for RNA clean-up prior to any downstream application. Purified RNA is suitable for mRNA isolation (Product R6511, mRNA Enrichment Kit), Northern analysis, RNase protection assays, differential display, and reverse transcription.
主要试剂
1. 2-mercaptoethanol ($-mercaptoethanol, $-ME) is required and must be added to TRK Lysis Buffer before use. Add 20 ul $-ME per 1 ml TRK Lysis Buffer. The mixture should be stored at room temperature and is stable for about 1 week.
2. 70% ethanol (in water) and absolute ethanol.
3. DEPC-treated deionized water.
主要设备
1. RNase-free 50 ml conical centrifuge tubes.
2. Centrifuge with swinging-bucket rotor at room temperature capable of 3,500 x g.
3. RNase-free pipet tips and disposable gloves.
4. Note that all centrifugation are performed using a centrifuge with a swingingbucket rotor at 4,000 -6,000 x g at ROOM TEMPERATURE.
实验步骤
Quantity of Starting Material: In general the binding capacity of the HiBind® RNA Maxi column (~5 mg RNA) will not be exceeded and between 0.1g and 1.0 g of tissue can be used. However, some tissues such as brain and skeletal muscle tend to be rather difficult to lyse and homogenize and can block the column, leading to lower RNA yields. With such tissues, we recommend starting with 250-400 mg to ensure optimal column performance. Subsequently, the starting quantity may be increased to 1.0g if favorable yield and quality is obtained. Overloading the column always leads to clogging and reduced yields.
Freezing Samples: Fresh tissue is best, but frozen samples may also be processed. To store tissues for future use, flash-freeze under liquid nitrogen and store at -70°C for 2-4 months. Frozen samples should be processed directly in TRK Lysis Buffer without prior thawing. Once homogenized in TRK Lysis Buffer, samples may be stored at - 70°C for several months.
1. In almost every case, optimal yields are only obtained if the tissue sample is first completely and thoroughly disrupted and homogenized in TRK Lysis Buffer. We suggest using a mechanical rotor homogenizer. Wearing gloves, place 250-400 mg tissue in a sterile RNase-free 50 ml polypropylene centrifuge tube (not provided). Add 7.0 ml TRK Lysis Buffer containing 2-mercaptoethanol and homogenize for 2 minutes.
Note: Remember to add 2-mercaptoethanol to TRK Lysis Buffer before use. Add 20 ul 2-mercaptoethanol per 1 ml TRK Lysis Buffer before use.
Tip: For difficult tissues, use up to 14.0 ml of TRK Lysis Buffer with no more than 0.4 g tissue and extend homogenization period.
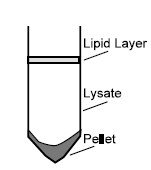
2. To obtain a cleared lysate centrifuge for 15 min at 5,000 x g. A loose pellet usually forms accompanied by an upper lipid layer.
3. Without disturbing the pellet or the lipid layer, carefully pipet only the cleared lysate to a fresh 50 ml polypropylene centrifuge tube (not provided).
Tip: Transferring contaminants from the pellet or the lipid layer will cause the spin column to clog resulting in significantly reduced yields. If necessary, sacrifice 1-3 ml lysate to ensure optimal yield and quality.
4. Add an equal volume (7.0 or 14.0 ml) of 70% ethanol to the cleared lysate from step 2. If some volume is lost in prior steps, add the appropriate amount of ethanol. Mix well by vortexing or vigorous shaking for 1 minute. Precipitates may form upon addition of ethanol. This will not interfere with the procedure so long as the mixture is thoroughly and immediately mixed. Proceed to step 5 immediately.
5. Apply sample to an HiBind® RNA maxi column assembled in a 50 ml collecting tube (provided). The spin column has a maximum capacity of approximately 25 ml. Cap the tube and centrifuge for 5 min at 5,000 x g at room temperature. If more than 0.4 g tissue was used, extend centrifugation time to 10-12 min. Discard flow-through liquid and reuse collecting tube in step 6.
Tip: If sample volume exceeds 25 ml, load the spin column in successive aliquots and centrifuge as above, discarding the flow-through liquid each time.
Optional: This is the starting point for on-membrane DNase I digestion treatment. See page 9 for detail protocol.
6. Add 15 ml RNA Wash Buffer I, cap the 50 ml collecting tube and centrifuge for 5 min at 5,000 x g at room temperature. Again discard flow-through liquid and reuse 50 ml tube in step 7.
7. Add 10 ml RNA Wash Buffer II diluted with absolute ethanol to the HiBind® RNA maxi column, cap the 50 ml tube and centrifuge for 3 min at 5,000 x g at room temperature. Discard flow-through liquid and reuse the 50 ml tube in step 8.
Note: RNA Wash Buffer II is supplied as a concentrate and must be diluted with absolute ethanol as described on page 4.
8. Repeat step 7 with another 10 ml RNA Wash Buffer II. Discard flow-through liquid and re-insert the spin column in the empty 50 ml collecting tube. Centrifuge the empty column for 15 min at 5,000 x g to dry the HiBind® matrix. This step is critical for removing traces of ethanol that may otherwise interfere with subsequent downstream applications.
9. RNA elution. Transfer the HiBind® RNA maxi column to a new 50 ml centrifuge tube (not supplied) and pipet 1-2 ml DEPC-treated water directly onto the matrix. For expected RNA yields greater than 1 mg use 1.25 ml of water. Cap the tube and allow the matrix to soak for 1 min. Centrifuge for 5 min at 5,000 x g.
Tip: Yield will be increased by 30%-70% by repeating the elution with a second volume of DEPC-treated water. For a higher final RNA concentration, this second elution may be performed using the first eluate. However, this will result in a ~30% lower overall yield.
Note: For laboratory centrifuges not capable of achieving 5,000 x g step 8 may not completely dry the column. As a result traces of ethanol may remain on the HiBind® RNA matrix and will be eluted with RNA. In this case it is best to further purify the RNA following elution by ethanol precipitation. Add 1/10 volume DEPC-treated 5 M ammonium acetate followed by 3 x vol absolute ethanol. In situations were expected RNA yields are less than 100ug or so, it is recommended to add 5 ug yeast tRNA or 10 ug glycogen as carrier prior to ethanol precipitation. Vortex to mix and incubate 2 hours at -70°C or overnight at -20°C. Centrifuge at 10,000 x g for 15 min at 4°C to pellet RNA. Aspirate supernatant and discard without disturbing pellet. Wash once with 75% ethanol and centrifuge as before. Discard supernatant and air-dry RNA pellet briefly before reconstituting in DECP-treated water.







