新的基因编辑领域突破口——表观遗传调控(二)
2. 神经系统疾病
▼ 致病机理:神经细胞中由于遗传缺陷导致的疾病
▼ 代表工作:同时另一项突破性的工作则使用一种SunTag(dCas9-10xGCN4)系统融合多个拷贝的转录激活蛋白(p65-HSF1),构建了一种Cre依赖性的SunTag-p65-HSF1(SPH)转基因小鼠模型。使用AAV8将Cre和sgRNAs递送到SPH转基因小鼠中,通过激活内源性神经源性转录因子的表达,在小鼠体内成功地实现了星形胶质细胞直接转化为功能性的神经元[17]。这一研究表明,在不需要使用外源重编程因子或转录因子的前提下,通过转录调控的CRISPR系统可以实现特定细胞的重编程或不同细胞类型之间的转分化,为体外细胞基因治疗遗传疾病提供了新的策略。
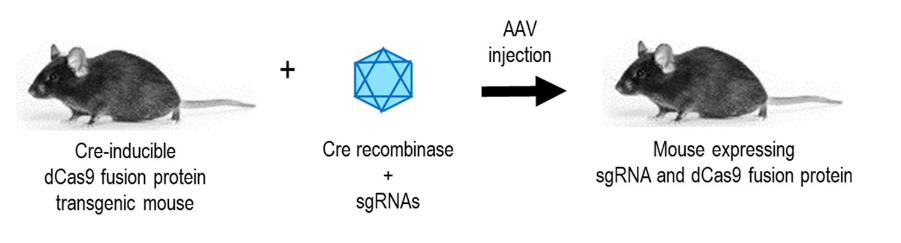
图5. Cre诱导的dCas9小鼠[21]
3. 单倍体剂量不足引起的疾病
▼ 致病机理:单倍剂量不足(haploinsufficiency )指一个等位基因突变后,另一个等位基因能正常表达,但这只有正常水平50%的蛋白质不足以维持细胞正常的生理功能。
▼ 代表工作:近日,Science在线发表了一篇使用CRISPRa系统在小鼠中成功修复一种因单倍剂量不足引起的肥胖。研究人员通过AAV在SIM1基因或MC4R基因部分功能丧失的小鼠脑部递送dCas9-vp64和sgRNA的方式成功激活了SIM1或MC4R蛋白的表达,成功抑制了肥胖的表型[18]。这一策略给罹患单倍体剂量不足引起的疾病患者带来转机。
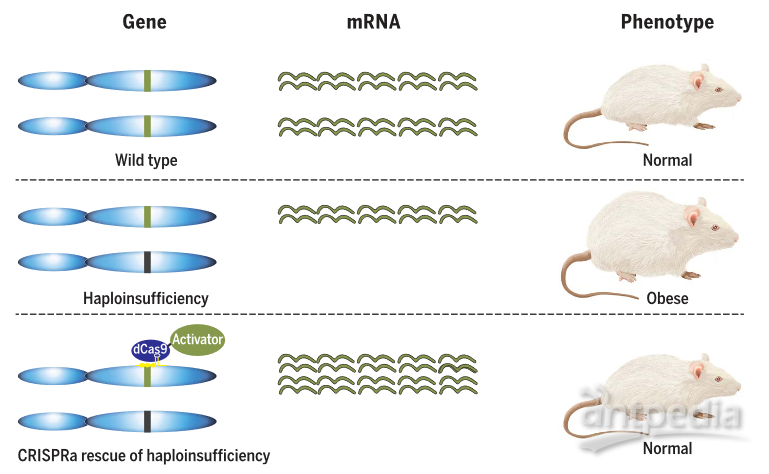
图6. 通过CRISPRa治疗单倍体剂量不足相关疾病[18]
4. 异常甲基化引起的疾病
▼ 致病机理:基因中CpG岛中的5' C经常突变引起高甲基化、羟甲基化等修饰,研究表明这些异常修饰会影响基因的表达调控,最终引起疾病[19]。
▼ 代表工作:脆性X综合征(Fragile X syndrome, FXS)就是一种由FMR1基因5' UTR 区中CGG三核甘酸重复序列扩增突变并高甲基化,使FMR基因沉默而导致的疾病。最近的一项研究通过利用dCas9融合Tet甲基胞嘧啶双加氧酶1(Tet methylcytosine dioxygenase 1, Tet1)转染FXS iPSCs 细胞系,成功靶向诱导FMR基因5' UTR CpG岛去甲基化,为这些因异常甲基化引起的疾病的治疗奠定了基础[20]。
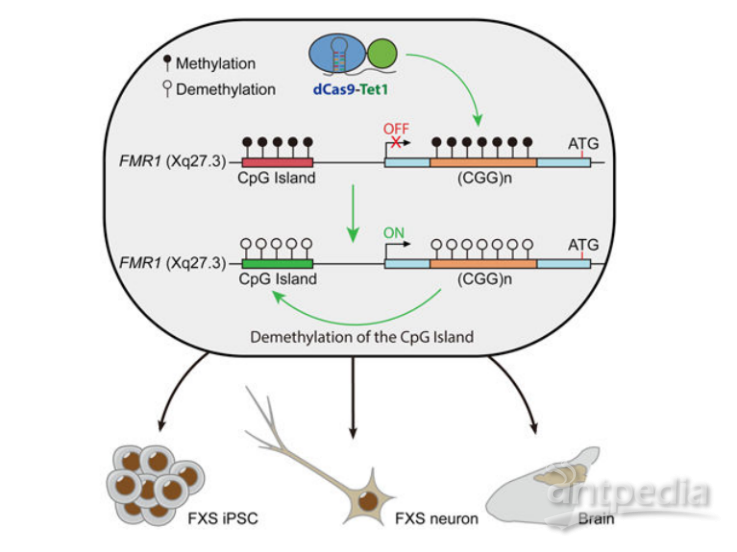
图7. 脆性X综合征相关疾病的表观遗传治疗[20]
怎么样,这次的公众号是不是让大家大开眼界呢?相信通过这四期的公众号,大家已经对基因编辑在疾病治疗中的研究应用有了较为系统的了解,不过“路漫漫其修远兮”,从实验室走到临床还有较为漫长的距离要走。但是大量的实验已经给我们看到了未来疾病治疗的新曙光,相信终有一天,基因编辑治疗的大时代终会到来。
参考文献:(向下滑动查看)
[1]Feinberg AP. The Key Role of Epigenetics in Human Disease Prevention and Mitigation. N Engl J Med. 2018, 378(14): 1323-1334.
[2]Brocken DJW, Tark-Dame M, Dame RT. dCas9: A Versatile Tool for Epigenome Editing. Curr Issues Mol Biol. 2018, 26:15-32.
[3]Thakore PI, D'Ippolito AM, Song L, Safi A, Shivakumar NK, Kabadi AM, Reddy TE, Crawford GE, Gers-bach CA. Highly specific epigenome editing by CRISPRCas9 repressors for silencing of distal regulatory ele-ments. Nat Methods, 2015, 12: 1143-1149.
[4]Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, Lim WA, Weissman JS, Qi LS. CRISPR-mediated modular RNA-guided regulation of tran-scription in eukaryotes. Cell, 2013, 154(2): 442-451.
[5]Maeder ML, Linder SJ, Cascio VM, Fu Y, Ho QH, Joung JK. CRISPR RNA-guided activation of endogenous human genes. Nat Methods, 2013, 10(10): 977-979.
[6]Perez-Pinera P, Kocak DD, Vockley CM, Adler AF, Kabadi AM, Polstein LR, Thakore PI, Glass KA, Oust-erout DG, Leong KW, Guilak F, Crawford GE, Reddy TE, Gersbach CA. RNA-guided gene activation by CRISPR-Cas9- based transcription factors. Nat Methods, 2013, 10(10): 973-976.
[7]Chavez A, Scheiman J, Vora S, Pruitt BW, Tuttle M, P R Iyer E, Lin S, Kiani S, Guzman CD, Wiegand DJ, Ter-Ovanesyan D, Braff JL, Davidsohn N, Housden BE, Perrimon N, Weiss R, Aach J, Collins JJ, Church GM. Highly efficient Cas9-mediated transcriptional programming. Nat Methods, 2015, 12(4): 326-328.
[8]Li Z, Zhang D, Xiong X, Yan B, Xie W, Sheen J, Li JF. A potent Cas9-derived gene activator for plant and mammalian cells. Nat Plants, 2017, 3(12): 930-936.
[9]Liu XS, Wu H, Ji X, Stelzer Y, Wu X, Czauderna S, Shu J, Dadon D, Young RA, Jaenisch R. Editing DNA methylation in the mammalian genome. Cell, 2016, 167(1): 233-247.
[10]Liu P, Chen M, Liu Y, Qi LS, Ding S. CRISPR-based chromatin remodeling of the endogenous Oct4 or Sox2 locus enables reprogramming to pluripotency. Cell Stem Cell, 2018, 22(2): 252-261.
[11]Cano-Rodriguez D, Gjaltema RA, Jilderda LJ, Jellema P, Dokter-Fokkens J, Ruiters MH, Rots MG. Writing of H3K4Me3 overcomes epigenetic silencing in a sustained but context-dependent manner. Nat Commun, 2016, 7: 12284.
[12]Kearns NA, Pham H, Tabak B, Genga RM, Silverstein NJ, Garber M, Maehr R. Functional annotation of native enhancers with a Cas9-histone demethylase fusion. Nat Methods, 2015, 12(5): 401-403.
[13]Hilton IB, D'Ippolito AM, Vockley CM, Thakore PI, Crawford GE, Reddy TE, Gersbach CA. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol, 2015, 33(5): 510-517.
[14]Kwon DY, Zhao YT, Lamonica JM, Zhou Z. Locus specific histone deacetylation using a synthetic CRISPRCas9-based HDAC. Nat Commun, 2017, 8: 15315.
[15]Mendenhall EM, Williamson KE, Reyon D, Zou JY, Ram O, Joung JK, Bernstein BE. Locus-specific editing of histone modifications at endogenous enhancers. Nat Biotechnol, 2013, 31(12): 1133-1136.
[16]Liao HK, Hatanaka F, Araoka T, Reddy P, Wu MZ, Sui Y, Yamauchi T, Sakurai M, O'Keefe DD, Núñez-Delicado E, Guillen P, Campistol JM, Wu CJ, Lu LF, Esteban CR, Izpisua Belmonte JC. In vivo target gene activation via CRISPR/Cas9-mediated trans-epigenetic modulation. Cell, 2017, 171(7): 1495-1507.
[17]Zhou H, Liu J, Zhou C, Gao N, Rao Z, Li H, Hu X, Li C, Yao X, Shen X, Sun Y, Wei Y, Liu F, Ying W, Zhang J, Tang C, Zhang X, Xu H, Shi L, Cheng L, Huang P, Yang H. In vivo simultaneous transcriptional activation of multiple genes in the brain using CRISPR-dCas9-activator transgenic mice. Nat Neurosci, 2018, 21(3): 40-446.
[18]Matharu N, Rattanasopha S, Tamura S, Maliskova L, Wang Y, Bernard A, Hardin A, Eckalbar WL, Vaisse C, Ahituv N. CRISPR-mediated activation of a promoter or enhancer rescues obesity caused by haploinsufficiency. Science, 2018.
[19]Xu Y, Wu F, Tan L, Kong L, Xiong L, Deng J, Barbera AJ, Zheng L, Zhang H, Huang S, Min J, Nicholson T, Chen T, Xu G, Shi Y, Zhang K, Shi YG. Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol Cell, 2011, 42(4): 451-464.
[20]Hayashi ML, Rao BS, Seo JS, Choi HS, Dolan BM, Choi SY, Chattarji S, Tonegawa S. Rescue of Fragile X Syndrome Neurons by DNA Methylation Editing of the FMR1 Gene. Cell, 2018, 172(5): 979-992.e6.
[21]Lau CH, Suh Y. In vivo epigenome editing and transcriptional modulation using CRISPR technology. Transgenic Res. 2018, 27(6):489-509.





















