Proteome 公司Scaffold 2.1软件现支持安捷伦Spectrum Mill 搜索平台
2008年8月7日安捷伦科技 分析测试百科 译
SANTA CLARA, Calif., and PORTLAND, Ore.,
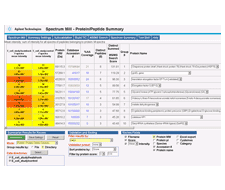
安捷伦科技和蛋白质组软件公司(Proteome Software)今天宣布,Proteome公司的Scaffold 2.1版本软件,现在可以处理从安捷伦的Spectrum Mill 蛋白数据库搜索引擎中搜索的结果。
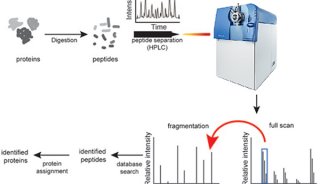
Scaffold可对串联质谱数据进行统计学认证,从多个搜索引擎中合并结果,并用图像显示在生物学相关的形态中的鉴定结果。Spectrum Mill 可对MS和MS/MS数据进行高通量的蛋白质数据库检索。
研究者可以把Spectrum Mill数据文件装载(load)到Scaffold中,然后把Spectrum Mill 的结果和各种搜索引擎结果比较,比如:Mascot,SEQUEST,Phenyx,OMSSA 和 X!Tandem。
“通过支持Spectrum Mill搜索引擎,Scaffold为它的集成库增加了另一个优秀的工具,使研究者们可以更容易地合并、比较、分享MS/MS检索结果。” Proteome公司的科学家Christopher Mason说。
“合并的搜索引擎数据,比任何单一的搜索引擎产生好得多的结果。”安捷伦实验室的研究者Alex Apffel博士说。“用户可以'拯救'那些可能会被拒绝的谱图;并为结果的评价提供了一个客观的统计基础。”
安捷伦实验室,该公司的中央研究机构同Proteome软件公司合作,集成了Spectrum Mill 和Scaffold,帮助研究者获得更高质量的数据。用于MassHunter工作站的Spectrum Mill 新版本,有几项独特的性能:
- 接受多数质谱仪器的数据;
- 是少数几个可以利用高质量精度MS和MS/MS谱图的软件之一,可以提高搜索结果的特异性和置信度
- 可以方便地鉴定未知的蛋白修饰;
- 提供优良的可视化工具,帮助自动和手动地验证、检查搜索结果,并显示差异的表达谱
若想下载Scaffold 2.1,请访问:www.proteomesoftware.com.
关于安捷伦的Spectrum Mill
MassHunter工作站上使用的Spectrum Mill,即使对于非常复杂的查询,反馈也很快速(~200 毫秒/张谱)。它通过快速的数据库搜索,独特的算法,智能并行和迭代搜索过程,快速地鉴定蛋白质和多肽。Spectrum Mill有多种操作模式:在鉴定模式下找到未修饰的肽段;在可变修饰或同源模式下可寻找翻译后修饰、变异或化学修饰。对于数据库中找不到的肽段,也可以进行从头(de novo)的谱图解析。Spectrum Mil可在无需同位素标记的情况下,鉴定两个数量级(或更高)的相对丰度差异;同时也支持所有的标记技术。
关于Proteome公司的软件
Scaffold 2.1是工业标准级的MS/MS后分析工具,对于串联质谱实验鉴定的蛋白,它给出一种完整的、实验性的展现。Scaffold帮助蛋白质组学研究人员验证,可视化,并分享他们的数据。更多关于Scaffold 2.1的信息,请访问:www.proteomesoftware.com
参考引文原文:
Agilent Technologies' Spectrum Mill Searches Now Supported in Proteome Software's Scaffold 2.1
[August 7, 2008 Agilent Technologies]
--------------------------------------------------------------------------------
SANTA CLARA, Calif., and PORTLAND, Ore., Aug. 6, 2008
Agilent Technologies and Proteome Software today announced that Proteome's Scaffold 2.1 software now supports the processing of search results from Agilent's Spectrum Mill protein database search engine.
Scaffold statistically validates peptides and proteins from tandem mass spectrometry data, combines results from multiple search engines and graphically displays identifications in biologically relevant formats. Spectrum Mill performs high-throughput protein database searches of MS and MS/MS data.
Researchers who load Spectrum Mill data files into Scaffold can now compare Spectrum Mill results with those of Mascot, SEQUEST, Phenyx, OMSSA and X!Tandem search engines.
"By supporting the Spectrum Mill search engine, Scaffold has added another excellent tool to its arsenal, making it even easier for researchers to combine, compare, and share MS/MS search results," noted Christopher Mason, staff scientist at Proteome Software.
"Combining search engine data yields a far better result than would be obtained with any one search engine," said Dr. Alex Apffel, Agilent Laboratories researcher. "Users can 'rescue' spectra that would otherwise be rejected and provide an objective statistical basis for evaluation of results."
Agilent Laboratories, the company's central research organization, collaborated with Proteome Software to help researchers achieve higher-quality data through the integration of Spectrum Mill and Scaffold. The new version of Spectrum Mill for MassHunter Workstation offers a number of unique capabilities:
Accepts data from most mass spectrometric instruments;
Is one of a few programs that exploit high mass accuracy data in MS and MS/MS spectra for improved specificity and confidence in search results;
Allows easy identification of unexpected protein modifications; and
Offers superior visualization tools to aid in automatic and manual validation, inspection of search results and display of differential expression.
To download Scaffold 2.1, please visit www.proteomesoftware.com.
About Agilent's Spectrum Mill
Spectrum Mill for MassHunter Workstation from Agilent Technologies provides rapid (~200 msec/spectrum) turnaround even for complex queries. It quickly identifies proteins and peptides via fast database searches, unique algorithms, intelligent parallelization and iterative search processes. Spectrum Mill can operate in identity mode to find unmodified peptides, or in variable modifications or homology mode to look for post-translational modifications, mutations, and chemical modifications. It also offers de novo spectral interpretation for peptides not found in any database. Spectrum Mill software can identify relative abundance differences of twofold or greater without complicated isotope labeling, but also supports all common labeling techniques.
About Proteome Software
Scaffold 2.1 Software is the industry-standard MS/MS meta-analysis tool that displays a complete, experiment-wide view of the proteins identified in a tandem mass spectrometry experiment. Scaffold helps proteomics researchers validate, visualize, and share their data. More information about Scaffold 2.1 is available on the web at www.proteomesoftware.com.