基于epMotion 5075t系统与KPPA HyperPlus试剂盒的全自动测序..1
基于epMotion 5075t系统与KPPA HyperPlus试剂盒的全自动测序前文库制备方案
Automated KAPA HyperPlus DNA Library Preparation for Illumina® Sequencing on the Eppendorf epMotion® 5075t
Nathan Quon1, Cheng Liu2, Ph.D., Eleanor Cowley1, M.S., Rachel Kasinskas1, Ph.D. and Dan Stover1, M.S.
1Roche Sequencing & Life Science | Kapa Biosystems, Wilmington, USA; 2Eppendorf AG, Hamburg, Germany
Abstract
The rapid growth and declining costs of Next Generation Sequencing (NGS)
have increased the demand for high throughput sequencing capacity.
Automation of library preparation offers customers efcient and effective
tools for meeting the increased throughput demands of the NGS world.
Eppendorf has partnered with Kapa Biosystems, an industry leader in life
science reagents, to develop an automated method for the KAPA HyperPlus
Library Preparation Kit on the Eppendorf epMotion 5075t automated
liquid handling system, as described in this application note.
Introduction
The KAPA HyperPlus Kit enables rapid construction of DNA libraries for
Illumina sequencing. It is compatible with a wide range of sample types
and input amounts (1 ng – 1 µg), making it one of the most versatile
kits on the market. The novel one-tube chemistry streamlines the DNA
fragmentation and library construction processes, yielding libraries of
similar or better quality than those produced with KAPA HyperPrep Kits
and Covaris®-sheared DNA. This highly optimized protocol, including
engineered enzymes, optimally formulated buffers, and minimal cleanup
steps, results in efcient conversion of input DNA to adapter-ligated
library, enabling deep and uniform sequence coverage.
The method developed on the Eppendorf epMotion 5075t offers an automated
solution for preparation of up to 48 sam ples, with the capability of
scaling up to 96 samples per run. Modular programming gives the user
flexibility to run a size selection step if desired, before or after PCR
amplifcation. The system’s user-friendly interface also guides the user
through the run setup, including placement of the labware and required
reagent volumes. An optical sensor verifes that all labware is correctly
placed before the run starts. Moreover, epMotion’s walk-away potential
for generating PCR-free libraries is maximized by an on-deck ThermoMixer
that provides homogeneous reaction mixtures, a thermal module that
enables on-deck incubation steps, and a gripper that transports plates
to various deck positions. Workflows with PCR require the use of an
off-deck thermocycler.

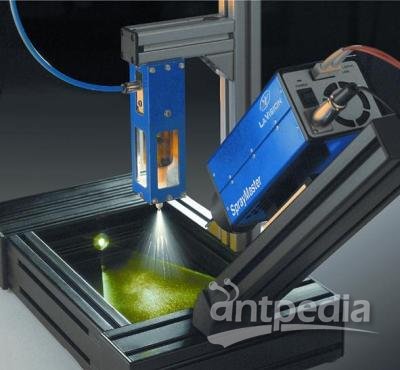
Figure 1: Eppendorf epMotion 5075t automated liquid handling system.
Materials and Methods
Experimental Design
Sixteen libraries were prepared from varying input amounts of
Escherichia coli (E.coli) genomic DNA to ensure the au tomated method
performed within the KAPA HyperPlus Kit specifcations. All incubations
were performed on-deck, with the exception of the library amplifcation.
Molecular biol ogy grade mineral oil, an inert overlay, was used to
prevent evaporation during the end repair and A-tailing incubation. Four
replicates each of 1 ng, 10 ng, 50 ng and 200 ng inputs were fragmented
at 37 °C for 35 minutes. End repair and A-tailing was performed at 65
°C for 30 minutes. The adapter concentrations were matched to the input
amounts accord ing to the kit specifcations, as shown below in Table 1.
The ligation reaction was performed at 20 °C for 15 minutes, followed by
a 0.8× post-ligation cleanup and 0.6× – 0.8× size selection. The number
of library amplifcation cycles was adjusted for each input amount to
achieve fnal library yields between 100 ng – 1 µg. The method was
completed with a fnal 1× post-amplifcation cleanup.

Table 1: Experimental summary of epMotion 5075t validation run. Sixteen libraries were prepared: four replicates each from 1 ng, 10 ng, 50 ng, and 200 ng input amounts with appropriate adapter concentrations and PCR cycles to generate between 100 ng – 1 µg of final library.
Quality control (QC) samples at several stages in the workflow were
collected during the validation of the automated method. QC samples were
recovered after the post-ligation cleanup, size selection, and the
post-amplifcation cleanup as shown in Figure 2. For each QC sample, 2 µL
of sample was stored in 18 µL of 10 mM Tris-HCl (pH 8.0 – 8.5) to
prevent degradation. Samples were quantifed using the KAPA Library
Quantifcation Kit (LQK), which measures library fragments containing the
sites necessary for flow cell hybridization and Illumina sequencing.
This quantifcation is a costeffective alternative to sequencing that can
indicate viability of the prepared libraries prior to downstream
sequencing.
Post-ligation and post-size selection QC samples were diluted 1:10,000
and post-amplifcation QC samples diluted 1:100,000 to ensure that all
data points fell within the standard curve of the KAPA LQK assay. All
qPCR samples and standards were run in triplicate (outlier data points
were excluded from the analyses) on the Eppendorf MasterCycler® thermal
cycling device. Final library size distributions were determined by
running 1:5 dilutions of the post-amplifcation libraries on the Agilent®
2100 BioAnalyzer® with the High Sensitivity DNA Assay.

Figure 2: KAPA HyperPlus workflow shown with optional QC checkpoints. The approximate total instrument time to run the 16 samples was 4 hours, including on-deck incubations. Size Selection, Library Amplifcation, and Post-Amplifcation Cleanup are optional steps that can be omitted, which would reduce the run time to ~2.5 hours.