Comprehensive identification of novel proteins and N-glycosylation sites四
Figure 2 Distribution of N-glycopeptides analyzed by different enriched methods and instruments of royal jelly proteins. A is the distribution of N-glycopeptides enriched by lectin and hydrazide methods using mass spectrometry of Q-Exactive (orbitrap-based MS). 21 and eight are N-glycopeptides uniquely identified by the lectin and hydrazide enrichment, respectively, and 18 are N-glycopeptides identified by both enrichment methods using orbitrap-based MS. B is the distribution of N-glycopeptides enriched by lectin and hydrazide methods using mass spectrometry of triple TOF 5600 (triple TOF-based MS). Eight and two are N-glycopeptides specifically identified by the lectin and hydrazide enrichment protocols, respectively, and six are N-glycopeptides identified by both nrichment methods using triple TOF-based MS. C is the distribution of N-glycopeptides identified by the orbitrap-based MS and triple TOF-based MS using lectin enrichment method. 29 are N-glycopeptides uniquely identified by orbitrap-based MS, and four are uniquely identified by triple TOF-based MS, and 10 are N-glycopeptides identified by both MS systems using the lectin enrichment method. D is the distribution of N-glycopeptides identified by orbitrap-based MS and triple TOF-based MS using hydrazide enrichment. 18 are N-glycopeptides identified by orbitrap-based MS alone, and eight are N-glycopeptides identified by both types of LC-MS/MS instruments with adoption of hydrazide enrichment.
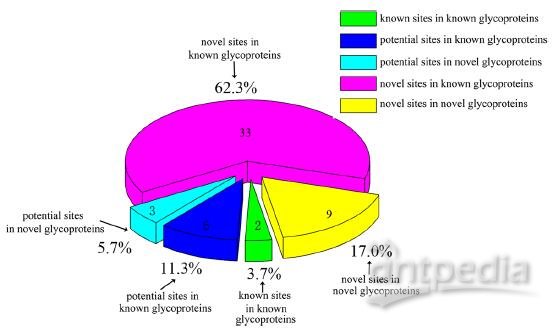
Figure 3 Distribution of N-glycosylated sites in royal jelly proteins. “2” is the identified two known sites in known glycoprotein. “6” is potential sites predicted in known glycoprotein, and “3” is potential glycosylation sites identified in novel glycoprotein. “33” is the novel sites identified in known glycoprotein, and “9” is the novel sites identified in novel glycoprotein.
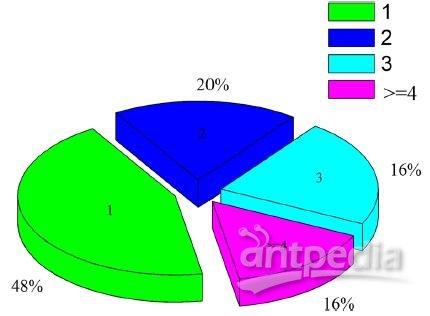
Figure 4 Distribution of N-glycosylated royal jelly proteins carrying different numbers of modification sites. “1, 2 and 3” are the N-glycosylated protein carrying 1, 2 and 3 N-linked glycosylation sites, respectvely. “> = 4” is the N-glycosylated protein carried four or more N-glycosylated sites.
As shown in Figure 3 and Table 2, the distribution of the 53 N-glycosylated sites was subdivided into known and novel proteins. Specifically, only two known sites in known glycoproteins were repeatedly identified in the current study, and six potential sites in known glycoproteins and three potential sites in novel glycoproteins were also identified. The potential sites predicted in the UniProt Database (updated April 2013) were also experimentally confirmed in this study. Thirty-three novel sites were identified in known glycoproteins, and nine novel sites in novel glycoproteins.
Site occupancy analyses showed that approximately 48% of N-glycosylated proteins carrying a single N-linked glycosylated site, 20% contained two sites, 16% retained three sites, and the rest carried four or more N-glycosylated sites (Figure 4).

To gain a better understanding the sequence motif of the N-linked glycosylation site in RJ, the surrounding sequences (five amino acids to both termini) of N-glycosylated sites were compared. As shown in Figure 5, about two-thirds were the N-X-T motif and the others were the N-X-S motif in the downstream (positive values) of N-linked modification sites. In other words, the N-linked sequence
motif was X-X-N-X-S/T-X in N-glycoproteins of RJ (N = asparagine, X = any amino acid except proline, S/T = serine or threonine).
Discussion
To gain a new understanding of innate biochemical properties of RJ at the proteome and glycoproteome levels, RJ was analyzed for the identification of novel proteins hidden in RJ and mapped for N-glycosylation sites using the double high LC-MS/MS system (orbitrap and triple TOF) and complementary methods of glycoprotein/glycopeptides enrichment (hydrazide chemistry and lectin). Overall, 13 novel proteins and 42 novel N-glycosylated sites in 25 N-glycosylated proteins were identified.
Identification of novel RJ proteins
The exploration of novel proteins in RJ is a long-term pursuit for apicultural biologists and biochemical experts. The fast improvement of MS with high resolution, high mass accuracy, and high sequencing speed now allows for in-depth identification of proteins in a comprehensive and unbiased manner in biological samples with high confidence. Compared with previous reports and bioinformatics analysis [1,11,17,28,29], 13 novel proteins were identified in this study. To establish the confidence that the newly identified proteins were real secretory proteins and not contaminated cellular proteins that may have leaked during secretory process of RJ glands, we used two bioinformatics software programs to confirm the origination of the secretory proteins. Proteins predicted as extracellular proteins by PSORT indicate they are putative secretory proteins [30]. To confirm this, SignalP was used to verify the presence of N-terminal secretory signal peptides [31]. This method suggested that all of the 13 novel proteins predicted to be secretory proteins are real protein components of RJ. They are mainly involved in metabolic processes and health promotion activities. This finding is of particular importance for opening new doors to understanding how RJ accomplishes its roles in honeybee biology and in the promotion of human health.
The YELLOW/MRJP is the most important RJ protein family and plays key roles both in honeybee biology and the promotion of human health [9]. The amazing fecundity of the queen (one queen lays 1,500-2,000 eggs a day, more than her body weight [2]) and the exponential speed of larval growth (weight increase by 1,600 times in the first six days of growth [32]) are achieved by a diet of highlynutritious RJ. MRJPs share a common evolutionary origin with the yellow protein family [33,34]. In particular, yellow- e3 and mrjp genes share the most introns/exons in the same relative positions [33]. The gene expression of yellow-e3 in the honeybee head and hypopharyngeal glands almost completely coincides with a developmental pattern typical of mrjp genes, supporting that yellow-e3 is the most recent common ancestor of the MRJP families [33,34]. Therefore, the newly identified yellow-e3 precursor in RJ is likely to act in a similar manner to that of the MRJPs, performing multifunctional roles in supplying nutrition and modulating caste determination of the honeybee [34,35]. Noticeably, in previous RJ studies, only MRJP1-5 have been repeatedly identified by a singular proteomics protocol [1,12,17,28]. MRJP6-9 are identified only when special technology is used [8,11]. For example identification of MRJP8 requires a special digestion method for the proteins [28]. In this study, we not only identified MRJP1-9 in a single study, but we also identified yellow-e3 precursor as a new member of the YELLOW/MRJP family. This indicates that our protocol has a high efficiency in identification of RJ proteins.







