Proteome and Phosphoproteome Quantification by DIA on Orbitrap (二)
FIGURE 2. Comparison of DDA and DIA raw data. (A) Comparison of MS1 base peak chromatograms. (B) Comparison of zoomedin chromatograms. (C) Composite score distribution of targets and decoys of DIA data.
DDA ID results were imported into Skyline as spectral library for DIA data processing. mProphet was used to perform statistical analysis of targets and decoys, and generated Q value for result refinement (Figure 2C). DDA IDs with q < 0.01 (Percolator) and DIA IDs with Q value < 0.01 were extracted as
high confident results.
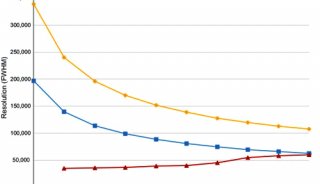
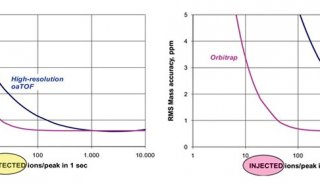
A total of 23072 peptides corresponding to 3794 proteins were identified from only 200 ng HeLa digest by DIA, showing good sensitivity of the ultra-high field Orbitrap. Moreover, 94.1% of the peptide IDs and 94.8% of the protein IDs were obtained from all three replicates of DIA, showing much higher reproducibility than DDA, as 57.0% of peptides and 72.8% of proteins identified from all replicates of DDA (Figure 3 & 4).
Conclusion
1) DIA has been applied and evaluated for large-scale protein and PTM quantification on the latest ultra-high field Orbitrap platforms.
2) A total of 21710 peptides from 3597 proteins were identified using DIA from only 200 ng HeLa digest, showing good sensitivity and reproducibility for quantification.
3) A total of 11341 phospho-peptides were identified using DIA and also displayed good sensitivity and reproducibility. Phospho-site ambiguity can be well distinguished, indicating the performance of DIA for PTM analysis.
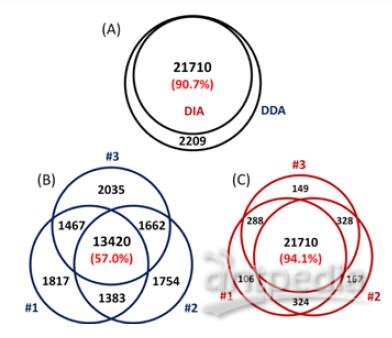
FIGURE 3. Sensitivity and reproducibility of peptide ID results of DDA and DIA. (A) More than 90% of library peptides (DDA result) can be identified from DIA with Q value of all three replicates <0.01. The ID reproducibility of DIA (C) is much better than DDA (B).
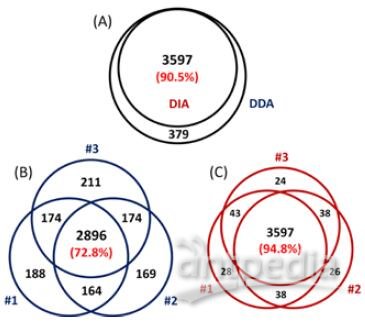
FIGURE 4. Sensitivity and reproducibility of protein ID results of DDA and DIA. (A) More than 90% of library proteins (DDA result) can be identified from all three replicates of DIA. The ID reproducibility of DIA (C) is also better than DDA (B).
The variations of transition/precursor peak areas were also calculated and compared to evaluate quantification ability of DDA and DIA. The CV of 95.2% DIA peptides are below or equal to 20%, while only 74.0% DDA peptides have a CV ≤ 20% (Figure 5).
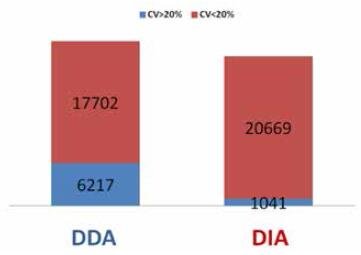
FIGURE 5. Comparison of peptide number with CV of transition/precursor peak area < 20% (red) or > 20% (blue). The quantification performance of DIA is much higher than DDA.
Data Independent Acquisition for Large- Scale Phosphorylation Analysis
The DIA strategy was further applied for phosphorylated protein sample to evaluate the performance of DIA for PTM analysis. A total of 11341 phospho-peptides were extracted with Q value <0.01 in all replicates. Figure 6 shows the DDA and DIA chromatograms as well as the DIA composite score distribution. Phospho-site ambiguity can be well distinguished due to high quality library spectra, good chromatogram separation and high sensitive DIA acquisition (Figure 7). DIA shows high ID reproducibility (Figure 8) and low peak area CV (Figure 9) as well in PTM analysis.
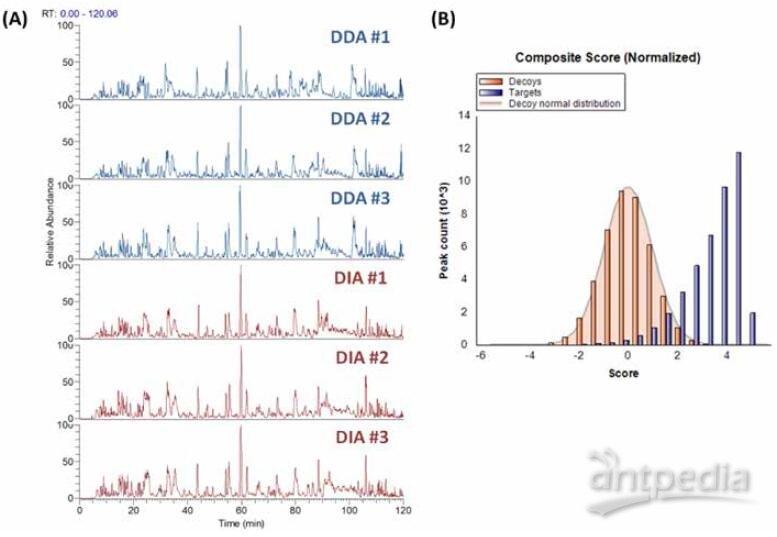
FIGURE 6. Comparison of phosphorylation sample data. (A) Comparison of MS1 base peak chromatogram. (B) Composite score distribution of targets and decoys of phosphorylation DIA data.
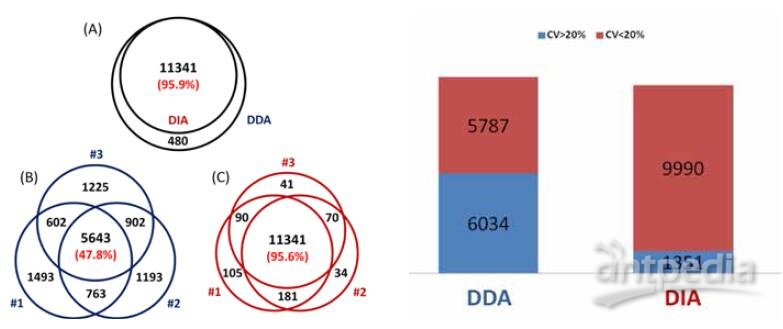
FIGURE 7. Sensitivity and reproducibility of phospho-peptide ID results. (A) More than 90% of library phospho-peptides (DDA result) can be identified from DIA with Q value of all three replicates < 0.01. The ID reproducibility of DIA (C) is much better than DDA (B).
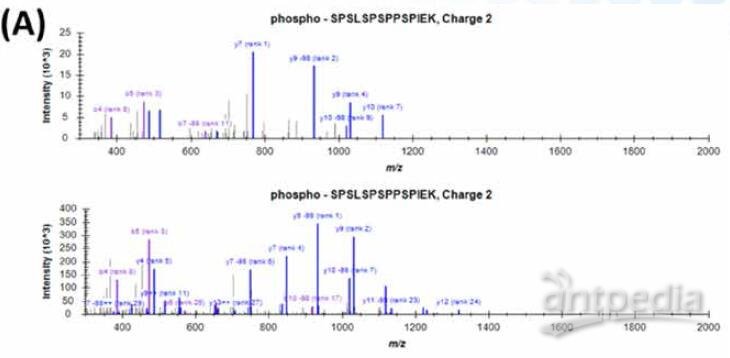
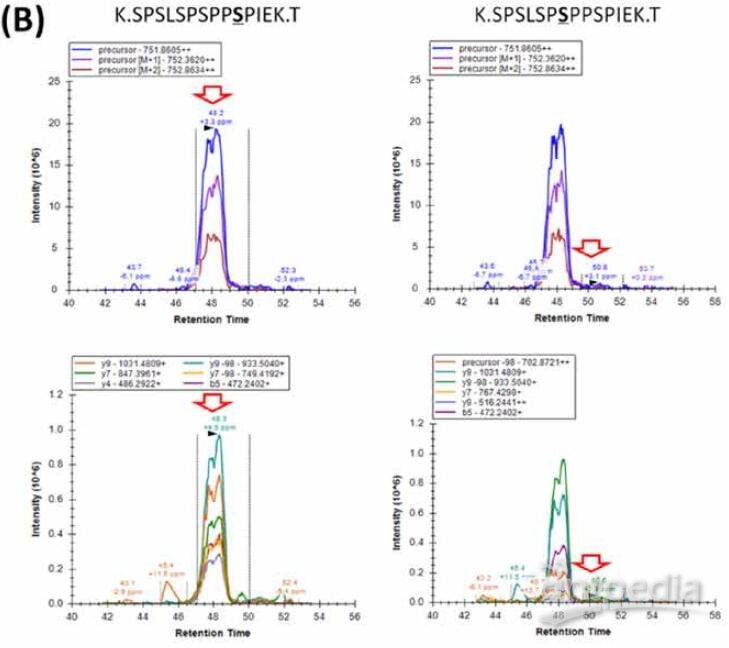
FIGURE 8. Example peptides with identical sequence and mass but different phosphorylation site. (A) Correct peak assignment was obtained due to high data quality.
-
企业风采

-
产品技术

-
焦点事件