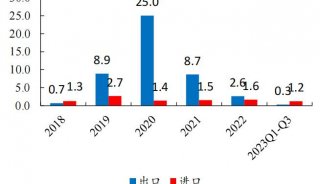
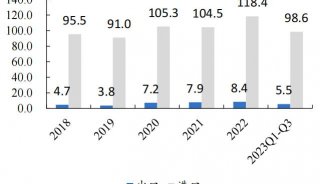
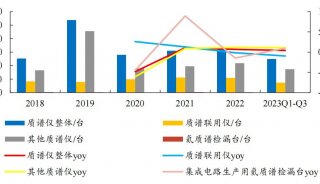
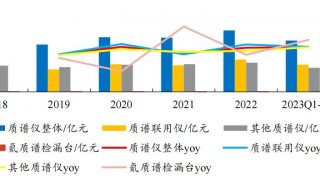
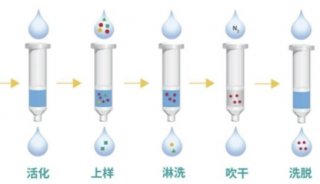
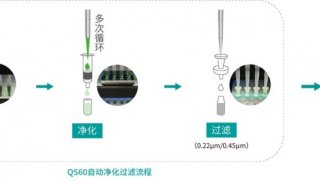
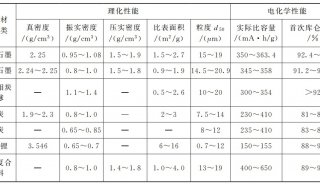
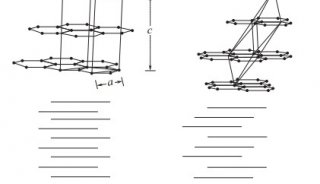
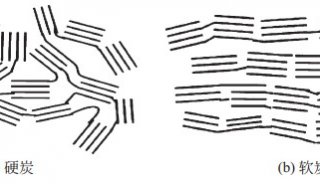
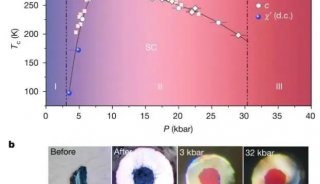
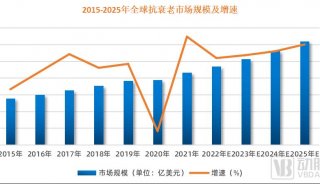
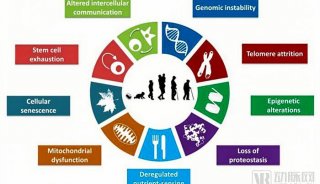
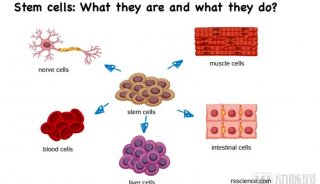
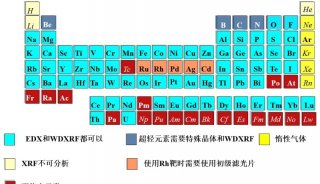
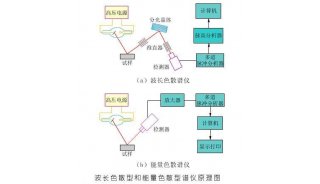
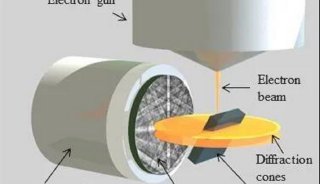
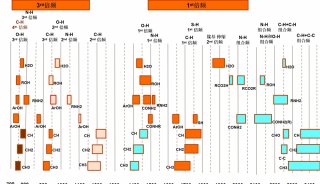
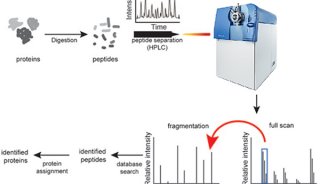
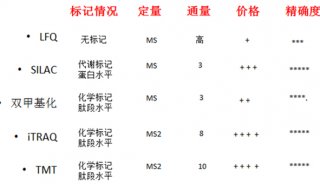
Immunoprecipitation... (一)
实验概要
We provide a general IP procedure including a list of reagents and a table to help you choose the correct protein beads.Immunoprecipitation is a method that enables the purification of a protein. An antibody for the protein of interest is incubated with a cell extract enabling the antibody to bind to the protein in solution. The antibody/antigen complex will then be pulled out of the sample using protein A/G-coupled agarose beads. This physically isolates the protein of interest from the rest of the sample. The sample can then be separated by SDS-PAGE for western blot analysis.
主要试剂
1. Lysis buffers
Salts: 0-1 M
Detergent, non-ionic: 0.1-2%
Detergent, ionic: 0.01-0.5%
Divalent cations: 0-10 mM
EDTA: 0-5 mM
pH: 6 – 9
2. Non-denaturing lysis buffer
20 mM Tris HCl pH 8
137 mM NaCl
1% Nonidet P-40 (NP-40)
2 mM EDTA
Store up to 6 months at 4°C. Immediately before use add protease inhibitors
3. RIPA (RadioImmunoPrecipitation Assay) buffer
50 mM Tris HCl pH 8
150 mM NaCl
1% NP-40
0.5% sodium deoxycholate
0.1% SDS
Store up to 6 months at 4°C. Immediately before use add protease inhibitors.
For convenience, a 10% sodium deoxycholate stock solution (5 g into 50 ml) may be prepared. It must be protected from light.
4. Detergent-free soluble protein lysis buffer
PBS containing:
5 mM EDTA
0.02 % Sodium Azide
Store up to 6 months at 4°C. Immediately before use add protease inhibitors
5. Denaturing lysis buffer/buffer for non-detergent soluble antigens:
1% SDS
5 mM EDTA
Store up to 1 week at room temperature
Immediately before use add:
10 mM dithiothreitol or beta-mercaptoethanol
Protease inhibitors
15 U/ml DNase1
6. Wash buffers
10mM Tris; adjust to pH 7.4
1mM EDTA
1mM EGTA; pH 8.0
150mM NaCl
1% Triton X-100
0.2mM sodium ortho-vanadate
Protease inhibitor cocktail
Store up to 6 months at 4°C. Immediately before use add protease inhibitors
7. Protease inhibitors
8. Other reagents required:
Sterile PBS pH 7.4
Sterile PBS-BSA 1% w/v (filtered)
TBST buffer
Loading/sample buffer for western blotting
100 mM EDTA stock solution is made with 1.86 g EDTA dissolved into 40 ml H2O. Add NaOH to adjust the pH to 7.4. Finally, adjust the total volume to 50 ml.
实验步骤
1. Preparation of lysates
1) Lysates from cell culture
Non denaturing:
a. Place the cell culture dish on ice and wash the cells with ice cold PBS.
b. Drain the PBS, then add ice cold lysis buffer (1 ml per 107 cells/100 mm2 dish/150 cm2 flask; 0.5 ml per 5x106cells/60 mm2 dish or 1875px2 flask).
c. Scrape adherent cells off the dish using a cold plastic cell scraper then gently transfer the cell suspension into a pre-cooled micro centrifuge tube.
d. Maintain constant agitation for 30 min at 4°C.
e. Centrifuge in a micro centrifuge at 4°C.
You may need to vary the centrifugation force and time depending on the cell type. A guideline is 20 min at 12,000 rpm but this must be determined by the end user (e.g. leukocytes need a very light centrifugation).
f. Gently remove the tubes from the centrifuge and place on ice. Aspirate the supernatant and place in a fresh tube kept on ice, and discard the pellet.
Denaturing:
a. Add 100 μl denaturing lysis buffer to 0.5 to 2 x 107 cells.
b. Mix well by vortexing vigorously for 2 to 3 sec at maximum speed. Transfer the cell suspension to a micro centrifuge tube.
The solution can be viscous at this stage due to release of DNA. |
c. Heat samples to 95°C for 5 min to denature.
d. Dilute the suspension with 0.9 ml non denaturing lysis buffer. Mix gently. (The excess 1% Triton X-100 in the non denaturing lysis buffer quenches the SDS in the original denaturing buffer).
e. Fragment the DNA by passing the lysed suspension 5 to 10 times through a needle attached to a 1 ml syringe.
Repeat mechanical disruption until the viscosity is reduced to manageable levels. If the DNA is not fully digested and fragmented, it can interfere with the separation of the pellet and supernatant following centrifugation. |
f. Incubate on ice for 5 min.
g. Proceed with the immunoprecipitation
2) Lysates from tissue
a. Dissect the tissue of interest with clean tools, on ice preferably, and as quickly as possible to prevent degradation by proteases.
b. Place the tissue in round bottom micro centrifuge tubes and immerse in liquid nitrogen to “snap freeze”. Store samples at -80°C for later use or keep on ice for immediate homogenization.
c. For a ~5 mg piece of tissue, add ~300 μl lysis buffer rapidly to the tube and homogenize with an electric homogenizer.
d. Rinse the blade twice with another 300 μl lysis buffer per rinse and then maintain constant agitation for 2 hr at 4°C (e.g. place on an orbital shaker in the refrigerator).
Volumes of lysis buffer must be determined in relation to the amount of tissue present. Protein extract should not be too dilute in order to avoid loss of protein and to minimize the volume of samples to be loaded onto gels. The minimum concentration is 0.1 mg/ml; optimal concentration is 1-5 mg/ml.
Note: If denatured samples are required, use denaturing lysis buffer and perform steps 2 to 5 from the denaturing protocol above. |
e. Centrifuge for 20 min at 12,000 rpm at 4°C in a micro centrifuge. Gently remove the tubes from the centrifuge and place on ice, aspirate the supernatant and place in a fresh tube kept on ice. Discard the pellet.
2. Pre-clearing the lysates
Pre-clearing the lysate can help reduce non specific binding of proteins to agarose or Sepharose beads. Pre-clearing with an irrelevant antibody or serum will remove proteins that bind immunoglobulins non-specifically. The end result will have a lower level of background and an improved signal to noise ratio. However, if the final detection of the protein is by western blotting, pre-clearing may not be necessary, unless a contaminating protein is interfering with visualization of the protein of interest.
1) Add either 50 μl of irrelevant antibody of the same species and isotype as the IP antibody or normal serum (rabbit is preferred by some researchers, see Harlow and Lane, page 243) to 1 ml of lysate. Incubate for 1 hr on ice.
2) Add 100 μl of bead slurry to the lysate.
3) Incubate for 10 to 30 min at 4°C with gentle agitation.
4) Spin in micro centrifuge at 14,000 x g at 4°C for 10 min.
5) Discard bead pellet and keep supernatant for immunoprecipitation.
To increase the yield, the beads can be washed 1 or 2 more times in lysis buffer, and the supernatants collected together.
It is important to make sure that as much of the normal serum is removed as possible as this will compete with the specific antibody against the antigen of interest. To check for this, a test can be done with lysis buffer instead of sample, performing all pre-clearing steps as above. A coomassie stain of a gel in which the resulting supernatant is run will reveal if the serum Ig is being removed effectively. If serum has not been sufficiently removed, bands will be present at 50 and 25 kDa for heavy and light chains; its presence may contribute to a weak IP. Consider either decreasing the amount of serum or increasing the amount of beads incubated with your samples in the pre-clearing
step.
-
并购

-
焦点事件

-
会议会展

-
焦点事件

-
企业风采

-
企业风采

-
焦点事件

-
焦点事件

-
焦点事件
-
焦点事件

-
企业风采

-
焦点事件
-
焦点事件

-
焦点事件

-
产品技术
-
焦点事件

-
焦点事件

-
焦点事件

-
焦点事件
-
科技前沿

-
科技前沿

-
焦点事件

-
企业风采

-
焦点事件

-
焦点事件

-
焦点事件

-
精英视角

-
焦点事件

-
技术原理
-
焦点事件

-
综述

-
精英视角

-
焦点事件

-
综述

-
焦点事件

-
焦点事件

-
焦点事件
-
焦点事件
-
焦点事件
-
焦点事件

-
焦点事件

-
焦点事件

-
焦点事件

-
焦点事件

-
焦点事件

-
焦点事件

-
焦点事件
-
焦点事件

-
焦点事件

-
焦点事件

-
焦点事件